Maize Cell Genomics: Resources for Visualizing Promoter Activity and Protein Dynamics Using Fluorescent Protein Lines
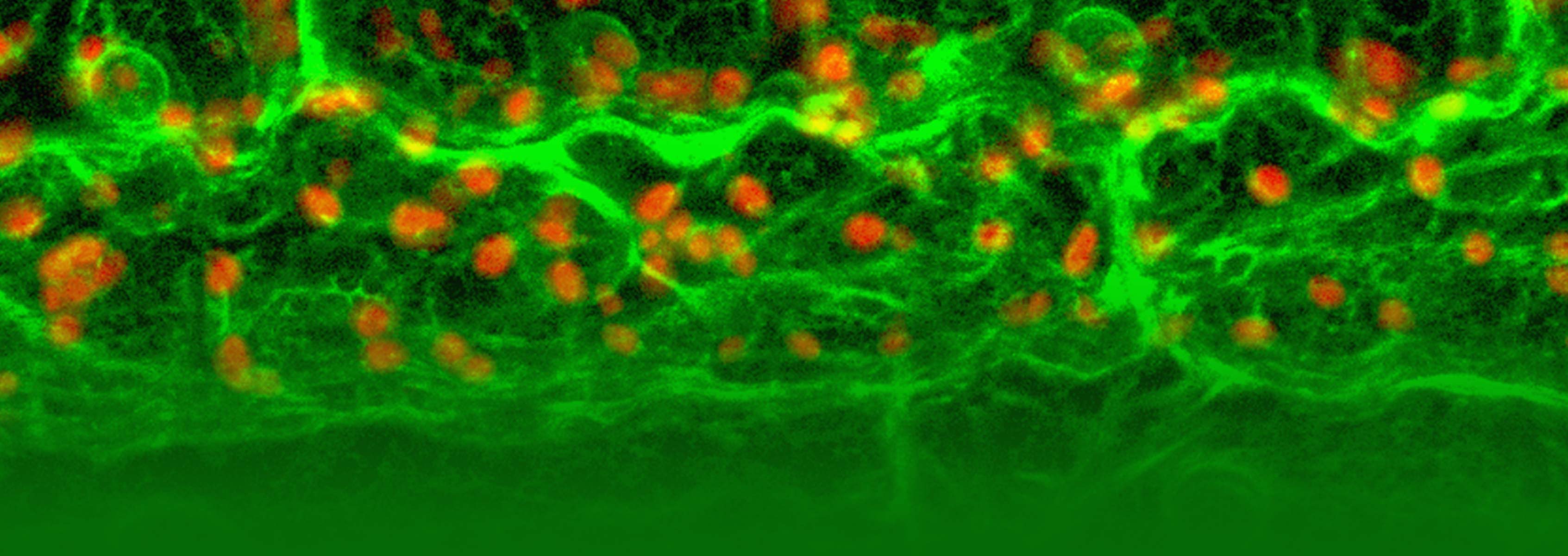
This NSF project has generated reporter lines that express proteins tagged with a fluorescent marker. Using confocal microscopy, the lines display visual information about when and where the tagged proteins are expressed and how specific proteins may be interacting and functioning. The new phase of the project will generate promoter fusion lines to drive expression of any gene in a number of specific chosen tissue or cell types, to enable gene over-expression and knockout studies.
Publications
Plant & cell physiology. 2015-01-01; 56.1: e12.
A maize database resource that captures tissue-specific and subcellular-localized gene expression, via fluorescent tags and confocal imaging (Maize Cell Genomics Database)
PMID: 25432973
Funding
Funding for thie project provided by National Science Foundation (NSF) awards #0501862 and #1027445.